Welcome to the Microbial Community Database (MiCoDa)
What is MiCoDa?
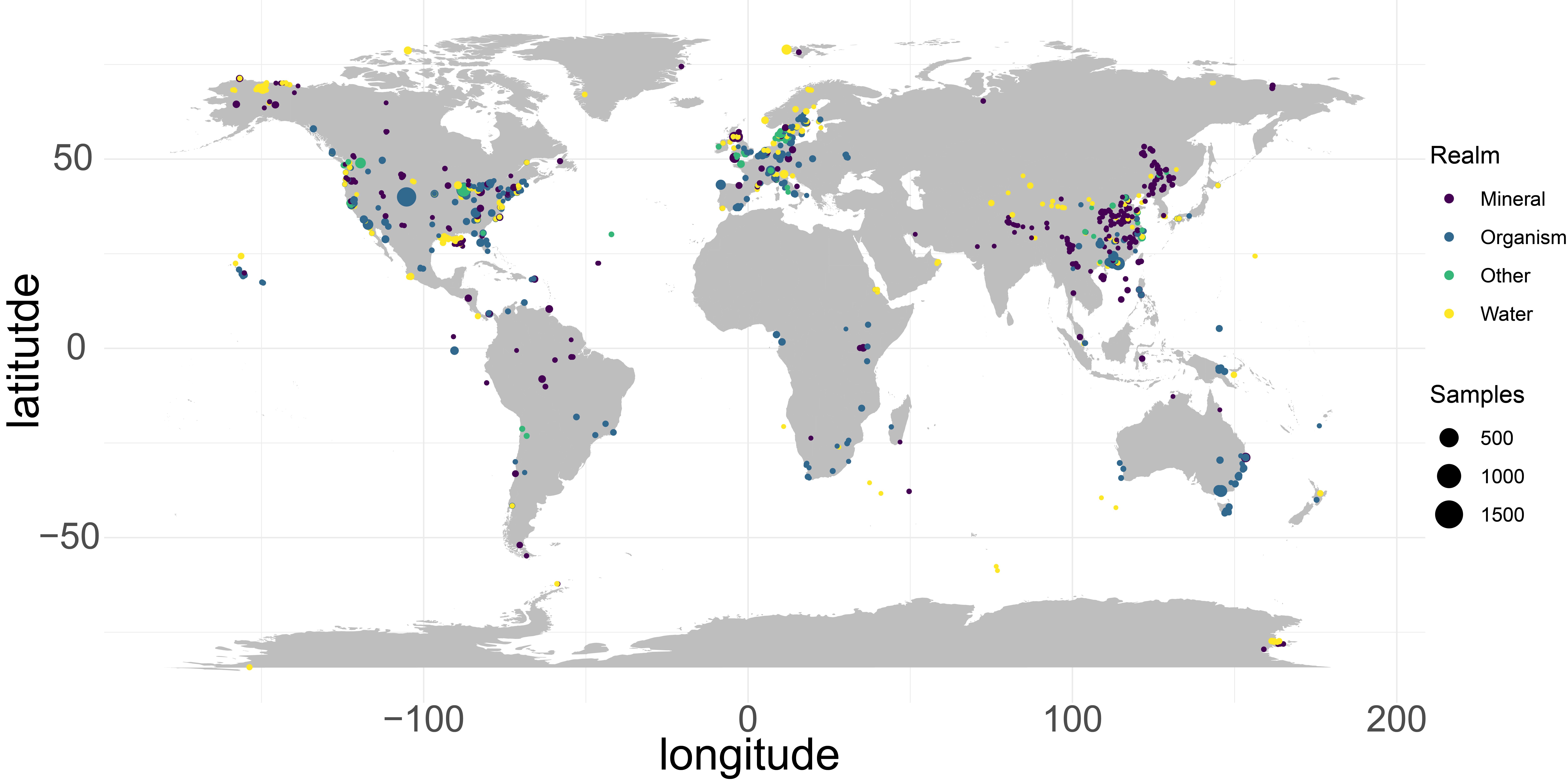
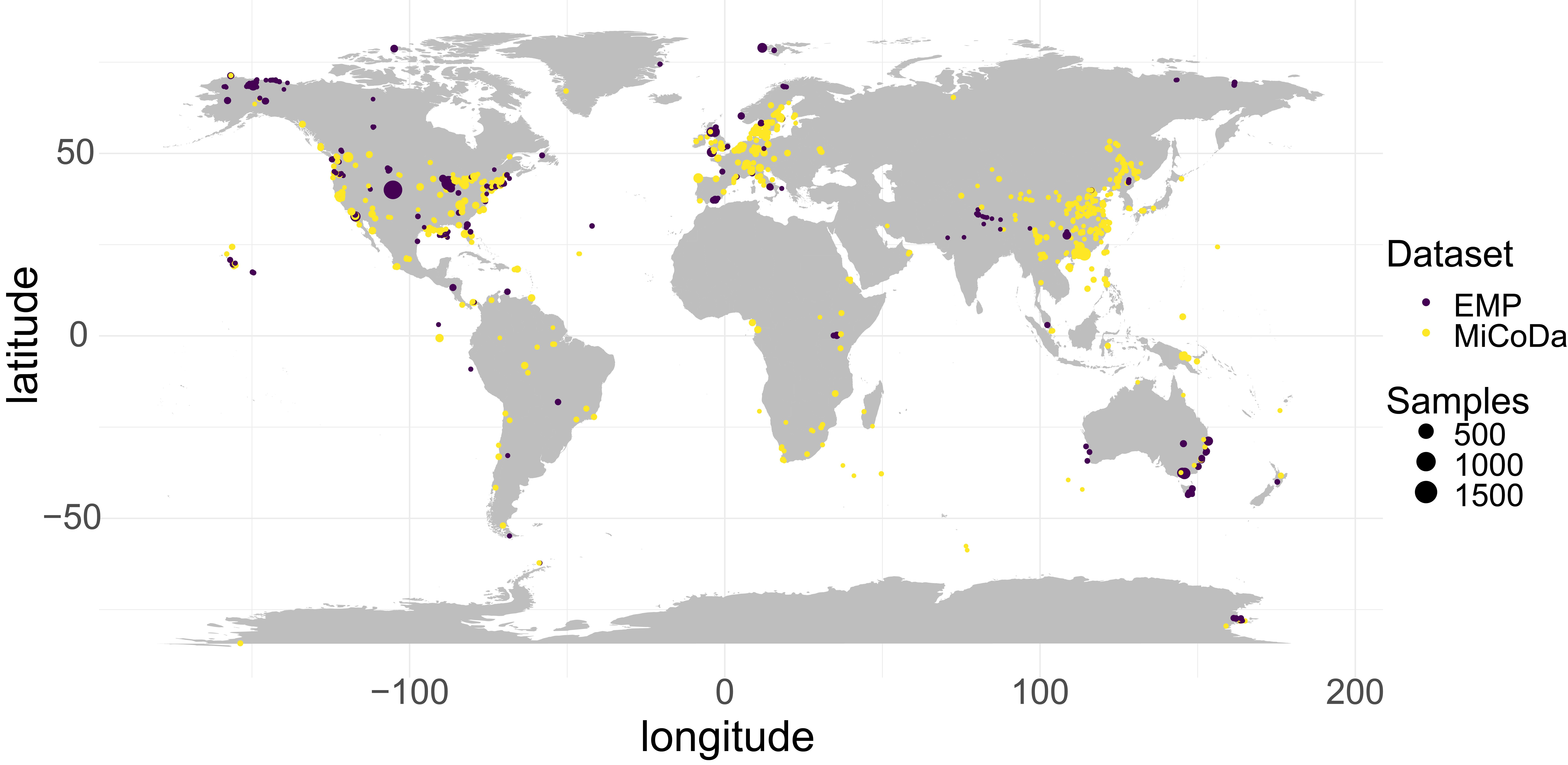
MiCoDa is a searchable database that hosts over 35,000 samples of processed 16S rRNA gene amplicon sequences from aquatic, host-associated, and mineral environments, spanning the entire globe. To improve cross-study comparability, all samples in MiCoDa have been sequenced in the same region of the 16S rRNA gene (between base pairs 515 and 806). MiCoDa also hosts the Earth Microbiome Project samples, processed in the same manner. MiCoDa is currently the largest public microbiome database available. Its goal is to encourage the reuse of extant sequence data by the life sciences. MiCoDa grew out of the observation that reusing biodiversity is hard, and it is especially complicated for microbiome sequence data. In addition to extensive data and metadata collection, the reuse of microbiome data requires extensive knowledge of bioinformatics and sufficient computational capacity for sequence processing. On the other hand, microbiome data is regularly archived. We created MiCoDa to take advantage of available data and facilitate microbiome data reuse and synthesis by specialists and non-specialists alike. To this end, we have manually curated the data and metadata included, preprocessed the sequence data to maximize comparability, and created a searchable data portal.
Who are we?
MiCoDa is a team effort led by Dr. Stephanie Jurburg (microbial ecology). It is hosted and supported by the Integrative Biodiversity Data and Code Unit of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, the Microbial Interaction Ecology group of the Helmholtz Centre for Environmental Research- Leipzig and the FUSION group of Friedrich Schiller Universität- Jena.
Who is MiCoDa for?
MiCoDa was designed for use by microbiome specialists and non-specialists. The ease of use encourages scientists to include bacteria in large-scale ecological syntheses, to place their novel microbiome findings within the context of extant data, to examine large scale patterns in microbiome data, etc. MiCoDa is also for data producers: the metadata files associated with MiCoDa samples include all the necessary bibliometric information to cite data producers, ensuring that they, too, receive credit.
Equitability in data reuse?
MiCoDa is designed to allow scientists to select samples based on relevant metadata (e.g., sample type or location) and download processed sequences in as .csv tables. Because the MiCoDa database does not require bioinformatics know-how or dedicated computing infrastructure, it is designed to increase worldwide scholarly access to microbiome datasets, reducing biases in who is able to perform large, comprehensive meta analyses, and facilitating equitability in synthesis research. Furthermore, because datasets are selected from mining published literature, original authors can be cited in later syntheses, increasing the visibility of extant research and giving credit to data providers. In its first version, MiCoDa nearly doubled the amount of microbiome data previously available with the EMP and greatly increased the geographic range and variety of available data. As part of its global data consolidation efforts, MiCoDa organizes yearly Datathon events. Find out more here.